6th
April 2018
"You should let the world know that the true source of mathematical methods as applicable to physics is to be found in the Proceedings of the Royal Society of Edinburgh. The volume- surface- and line- integrals of vectors and quaternions and their properties as in the course of being worked out by Tait is worth all that is going on in other seats of learning."
- James Clerk Maxwell, 1871.
In this blog we discuss the application of the knot theory in DNA structures. We will continue our discussions in Quantum Computing series afterwards. We had earlier described the importance of the measurement capacity of a macroscopic observer in j-space and how it results in an Unknot (Shallow Well) and Trefoil knot (Box). We next developed the description of a Trefoil knot in j-space. The description was in the form of Laurent polynomials and it satisfied the following properties:
- The structures for each knot-orientation were clearly distinguishable from each other. Each polynomial representation was unique.
- Symmetries corresponding to Charge (state and anti-state, 'h'), Parity (reflection along an axis or the equivalently ACW or CW knot orientation) and Time (order of the crossings, 'h'), were addressed within the same polynomial.
- Chiral property could be distinguished.
The Trefoil knots represented the structures as measured by the maximum efficiency observer Obsc (v~c). The measurements made by the next most efficient observer is represented by the Figure-8 knot. We explain further the description for the Figure-8 knot in j-space. Afterwards we will discuss a rather interesting application of the knot theory in unknotting of DNA.
The knot diagram for the
figure-8 knot, is shown as following:
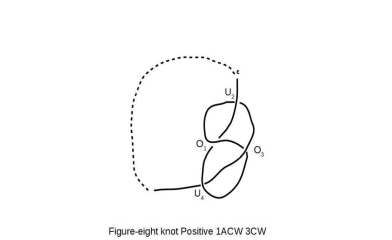
The knot polynomial for the figure-8 knot can be written, in the manner similar to the one introduced for Trefoil knot earlier as following:
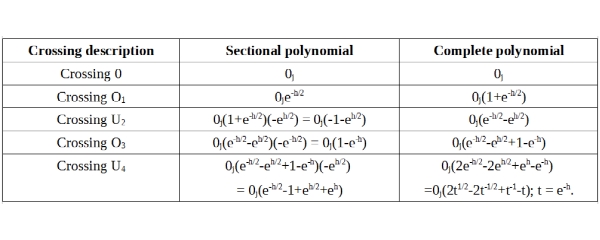
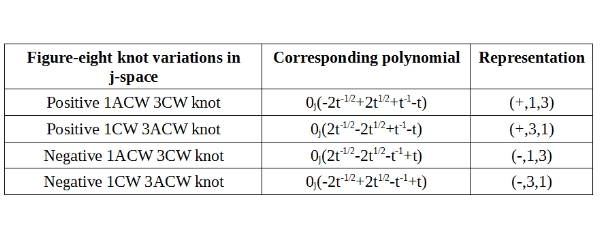
If we compare the Trefoil
and Figure-8 knots, we notice the absence
of constant in the knot polynomial for
Figure-8 knot. The Trefoil knot in
contrast has for example, a value "3"
representing the contributions of 0j.
This is an important difference as the
absence of the contribution of 0j
in the knot polynomial for figure-8 knot,
represents a corresponding physical
structure, which has no memory of the
initial state.
The structures with no
memory of the initial state,
are statistical in nature described
by the random variables. Thus if we
write a knot polynomial which shows no
contribution of 0j,
then it is likely to be of statistical
nature. Such structures can be further
reduced by a process similar to symmetry
breaking, until contribution of 0j is accounted for, in
the knot polynomials. The most
fundamental structure in j-space is
represented by Trefoil knot, which has
three alternating crossings (the required
minimum), to form a stable knot.
A Comparison Criterion for DNA and Enzymes:
A fantastic application will be to apply j-space knot polynomials, to the mechanism used by enzymes to unpack DNAs for replication. In j-space, enzymes and DNA both are observers, who are making measurements and being measured at the same time. Each molecule in a DNA represents a measurement. Subsequently by stringing together these measurements, a knot polynomial can be written for a specific DNA.
The DNA represented by knot polynomials with no contribution from 0j term, are more likely to be affected by enzymes and hence easy to replicate. If the enzyme is related to a potentially catastrophic disease then such DNA are at risk. Similarly the DNA represented by very high contributions of 0j term in their knot polynomials, are likely to be much more robust.
DNA and Enzymes
Taking the
argument further we can possibly also
write similar knot polynomials for
enzymes in j-space. If we compare
the knot polynomials written for enzymes
and DNA, the enzymes corresponding to
the polynomials of lower order (t-n,
t+n) and with high
contributions of 0j terms,
are more likely to succeed in breaking
down DNA with higher order polynomials
which have small contributions of 0j
terms.
In similar fashion, the DNA with lower order polynomials and with high contributions from 0j terms, should be immune to the effects of enzymes corresponding to higher order polynomials and with little contributions from 0j terms.
In similar fashion, the DNA with lower order polynomials and with high contributions from 0j terms, should be immune to the effects of enzymes corresponding to higher order polynomials and with little contributions from 0j terms.
***
Previous Blogs:
Chiral Symmetry
Sigma-z and I
Spin Matrices
Rationale behind Irrational Numbers
The Ubiquitous z-Axis
Majorana
ZFC Axioms
Set Theory
Nutshell-2014
Knots in j-Space
Supercolliders
Force
Riemann Hypothesis
Andromeda Nebula
Infinite Fulcrum
Cauchy and Gaussian Distributions
Discrete Space, b-Field & Lower Mass Bound
Incompleteness II
The Supersymmetry
The Cat in Box
The Initial State and Symmetries
Incompleteness I
Discrete Measurement Space
The Frog in Well
Visual Complex Analysis
The Einstein Theory of Relativity
***
Sigma-z and I
Spin Matrices
Rationale behind Irrational Numbers
The Ubiquitous z-Axis
Majorana
ZFC Axioms
Set Theory
Nutshell-2014
Knots in j-Space
Supercolliders
Force
Riemann Hypothesis
Andromeda Nebula
Infinite Fulcrum
Cauchy and Gaussian Distributions
Discrete Space, b-Field & Lower Mass Bound
Incompleteness II
The Supersymmetry
The Cat in Box
The Initial State and Symmetries
Incompleteness I
Discrete Measurement Space
The Frog in Well
Visual Complex Analysis
The Einstein Theory of Relativity
***
Attribution — You must give appropriate credit, provide a link to the license, and indicate if changes were made. You may do so in any reasonable manner, but not in any way that suggests the licensor endorses you or your use. No additional restrictions — You may not apply legal terms or technological measures that legally restrict others from doing anything the license permits. This is a human-readable summary of (and not a substitute for) the license.

